Repo is a data-centered data flow manager. It allows to store R data files in a central local repository, together with tags, annotations, provenance and dependence information. Any saved object can then be easily located and loaded through the repo interface.
A paper about Repo has been published in BMC Bioinformatics.
Latest news are found in the NEWS.md file of the “Untested” branch.
Creating a dummy repository under the R temporary folder (skipping confirmation):
library(repo)
rp <- repo_open(tempdir(), force=T)
#> Repo created.Storing data. In this case, just item values and names are specified:
God <- Inf
rp$put(God) ## item name inferred from variable name
rp$put(0, "user") ## item name specifiedMore data with specified dependencies:
rp$put(pi, "The Pi costant", depends="God")
rp$put(1:10, "r", depends="user")Loading items from the repository on the fly using names:
diam <- 2 * rp$get("r")
circum <- 2 * rp$get("The Pi costant") * rp$get("r")
area <- rp$get("The Pi costant") * rp$get("r") ^ 2Storing more data with verbose descriptions:
rp$put(diam, "diameters", "These are the diameters", depends = "r")
rp$put(circum, "circumferences", "These are the circumferences",
depends = c("The Pi costant", "r"))
rp$put(area, "areas", "These are the areas",
depends = c("The Pi costant", "r"))Showing repository contents:
print(rp)
#> ID Dims Size
#> God 1 51 B
#> user 1 49 B
#> The Pi costant 1 55 B
#> r 10 99 B
#> diameters 10 75 B
#> circumferences 10 103 B
#> areas 10 103 Brp$info()
#> Root: /tmp/RtmppFMwnb
#> Number of items: 7
#> Total size: 535 Brp$info("areas")
#> ID: areas
#> Description: These are the areas
#> Tags:
#> Dimensions: 10
#> Timestamp: 2019-12-22 17:01:45
#> Size on disk: 103 B
#> Provenance:
#> Attached to: -
#> Stored in: /tmp/RtmppFMwnb/a/areas
#> MD5 checksum: 56ad410055fedb0cae012d813a130291
#> URL: -Visualizing dependencies:
rp$dependencies()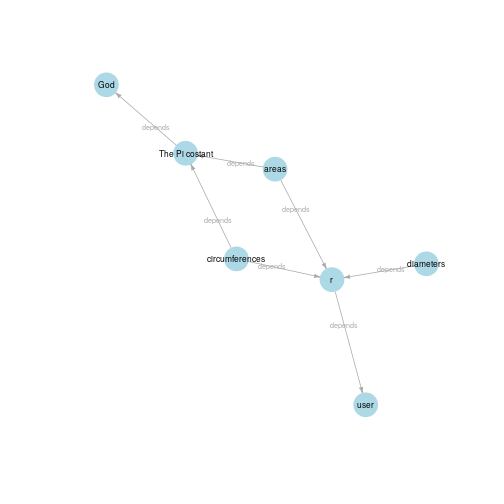
Manual acces to stored data:
fpath <- rp$attr("r", "path")
readRDS(fpath)
#> [1] 1 2 3 4 5 6 7 8 9 10Master: stable major releases, usually in sync with the latest CRAN version.
Dev: minor releases passing automatic checks.
Untested: in progress versions and prototype code, not necessarily working.
Besides inline help, two documents are available as introductory material:
Repo is on CRAN and can be installed from within R as follows:
install.packages("repo")Latest stable release can be downloaded from Github at https://github.com/franapoli/repo. Repo can then be installed from the downloaded sources as follows:
install.packages("path-to-downloaded-source", repos=NULL)devtools users can install Repo directly from github as
follows:
install_github("franapoli/repo", ref="dev")