

This package computes confidence intervals based on profile
log-likelihood for one or more parameters in a user-supplied fitted
multi-parameter model. The functionality of the main function,
profileCI(), is like that of confint.glm,
which calculates confidence intervals for the parameters of a
Generalised Linear Model (GLM).
Speed of computation can be improved by starting the profiling from limits based on large sample normal theory. The accuracy of the limits can be set by the user. A plot method visualises the log-likelihood and confidence interval. Cases where the profile log-likelihood flattens above the value at which a confidence limit is defined can be handled, leading to a limit at plus or minus infinity. Disjoint confidence intervals will not be found.
We illustrate the use of profileCI() using an example
from the help file for stats::glm().
## From example(glm)
counts <- c(18, 17, 15, 20, 10, 20, 25, 13, 12)
outcome <- gl(3, 1, 9)
treatment <- gl(3, 3)
glm.D93 <- glm(counts ~ outcome + treatment, family = poisson())
# Intervals based on profile log-likelihood
confint(glm.D93)
#> Waiting for profiling to be done...
#> 2.5 % 97.5 %
#> (Intercept) 2.6958215 3.36655581
#> outcome2 -0.8577018 -0.06255840
#> outcome3 -0.6753696 0.08244089
#> treatment2 -0.3932548 0.39325483
#> treatment3 -0.3932548 0.39325483To calculate these intervals using profileCI we provide
a function that calculates the log-likelihood for this Poisson GLM for
an input parameter vector pars.
poisson_loglik <- function(pars) {
lambda <- exp(model.matrix(glm.D93) %*% pars)
loglik <- stats::dpois(x = glm.D93$y, lambda = lambda, log = TRUE)
return(sum(loglik))
}The function profileCI() profiles the log-likelihood,
with respect to one parameter at a time. For a given value of this
parameter the log-likelihood is maximised over the other parameters. The
aim is to search below and above the MLE of the parameter until the
profile log-likelihood drops to a level corresponding to the limits of
the confidence interval of a desired confidence level.
Two arguments can be used to affect the speed with which the
confidence limits are obtained: mult determines the amount,
as a percentage of the estimated standard error of the estimator of the
parameter of interest, by which the value of the parameter is
incremented when profiling. Larger values of mult should
result in a faster calculation but increase the risk that one of the
optimisations required will fail. If the argument
faster = TRUE then the searches for the lower and upper
confidence limits are started from limits based on the approximate large
sample normal distribution for the maximum likelihood estimator of a
parameter, rather than the maximum likelihood estimate. The defaults are
mult = 32 and faster = TRUE.
library(profileCI)
prof <- profileCI(glm.D93, loglik = poisson_loglik)
prof
#> 2.5% 97.5%
#> (Intercept) 2.6958271 3.36656379
#> outcome2 -0.8576884 -0.06255514
#> outcome3 -0.6753594 0.08244109
#> treatment2 -0.3932489 0.39324886
#> treatment3 -0.3932489 0.39324886We can visualise the profile likelihood for a parameter using a plot method.
plot(prof, parm = "outcome2")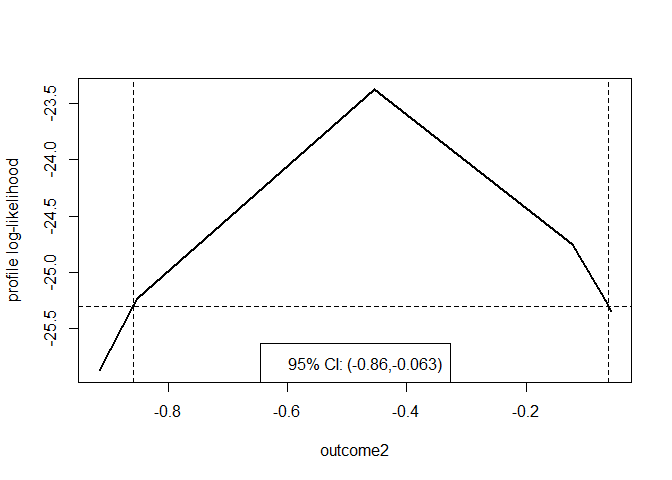
To obtain smooth version of this plot, we call
profileCI() with mult = 8 and
faster = FALSE, but this is a much slower calculation.
prof <- profileCI(glm.D93, loglik = poisson_loglik, mult = 8, faster = FALSE)
plot(prof, parm = "outcome2")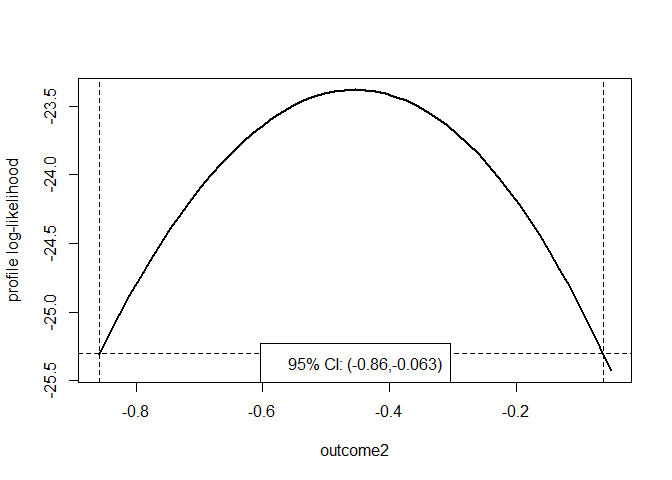
By default, once it has been determined that a limit lies between two
values of the parameter, quadratic interpolation is used to estimate the
value of the limit. If a specific degree of accuracy is required then
this can be set by passing a positive tolerance epsilon to
the itp function in the itp package.
An alternative to passing the log-likelihood function using the
argument loglik is to provide the same function via a
logLikFn S3 method for the fitted model object.
To get the current released version from CRAN:
install.packages("profileCI")To install the development version from GitHub:
remotes::install_github("paulnorthrop/profileCI")